Maize ectopic TF expression atlas
As part of NSF Award #IOS-2315723, we are surveying the response to expressing a large collection of cloned maize TFs in leaf protoplasts. We have established high-throughput assays to transform hundreds of TFs in parallel using robotics, and then assess the full transcriptome-wide response to each by RNA-seq. TFs included in the primary selection are from the Grassius TFome collection (https://grassius.org/tfomecollection). We are open to including additional TFs of interest to the community! See below if you are interested in contributing your favorite TF to this project.
Project Team: UGA – Brad Nelms (PI), Wayne Parrot (Co-PI), Taylor Strayhorn (PhD Student), Sungjin Park (Research Scientist); NC State – Cranos Williams (Co-PI), Teague McCracken (PhD Student)
Q: What is the ectopic TF expression atlas?
Why ectopic expression? By ectopically expressing TFs at new times and places, it is possible to induce their target pathways and reprogram plant development. For example, two TFs, Wuschel2 (Wus2) and Baby boom (Bbm), stimulate regeneration from transformed genotypes of maize, rice, and barley that were historically recalcitrant, overcoming a major bottleneck in cereal crop transformation. Ectopic expression can also give clues about native TF function, providing complementary information to genome-wide binding assays (e.g. DAP-seq) and loss-of-function genetics.
We have established methods for high-throughput transformation of maize leaf protoplasts in 384-well plates. Median transformation efficiencies are >70% and consistent between days:
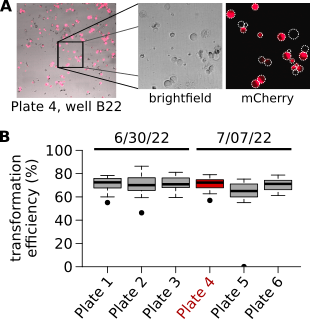
After transformation, we measure the transcriptome by RNA-seq and have found that expressing individual TFs is sufficient to induce distinct and reproducible responses in protoplasts:
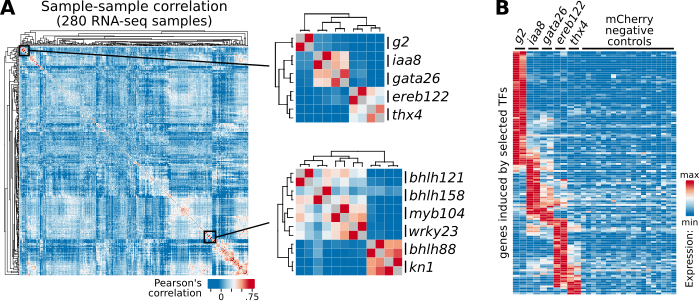
We will apply these assays to the full maize TFome (~2000 TFs and other DNA regulatory genes, see https://grassius.org/tfomecollection). These data will provide a valuable resource to the plant community and be made accessible via SRA and MaizeGDB.
Q: Could you include my favorite TF(s) in your study?
Yes! We are open to including additional maize TFs that are not part of the TFome. Please contact Brad Nelms for details (nelms@uga.edu).
Q: Where can I find your protoplast transformation protocols?
We use maize seedling leaf mesophyll cells for protoplast transformation. Our protocol is loosely based on Cao, et al. (2014). The pdfs below contain our working protocols for tube- and plate-based transformation as well as our modified DNA miniprep protocol to produce higher yields of DNA. We plan to publish a more thorough version of our plate-based transformation in the future.
