Research
We study pollen development using a blend of classic techniques in genetics and microscopy with powerful new genomic tools, such as single-cell RNA-sequencing. Current projects include:
Deciphering and controlling cell fate in plants
What if it was possible to reprogram a seedling leaf cell directly into pollen? This would shortcut a large portion of a plant’s life cycle as well as make key genetic events during meiosis accessible to manipulation. We are dissecting the gene regulatory circuits that control cell fate in plants, with an emphasis on pollen development. By combining research into the pathways that control pollen differentiation in vivo with synthetic approaches to reprogram cell fate in vitro, our aim is to both gain a deeper understanding of cell differentiation and establish new tools in plant breeding.
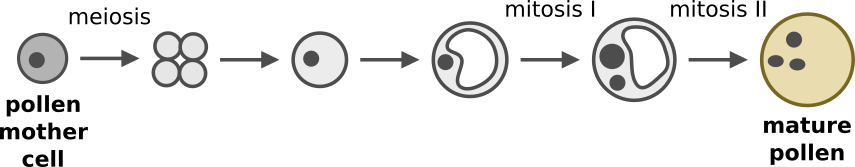
Transcriptional regulation during pollen development. Maize pollen makes a great model of cell differentiation because pollen precursors can be readily isolated at defined stages for genomics and biochemistry. Previously, we applied single-cell RNA-sequencing to identify several periods of major transcriptional change during pollen development (link). We have identified candidate genes that might control gene expression during these transitions, and will next evaluate the role of these genes in development.
Reprogramming the plant cell. Richard Feynman once said: “What I cannot create, I do not understand.” In the context of cell differentiation, a key test of understanding is the ability to transform a cell from one fate to another. A wide variety of cell lineage transformations have been achieved by ectopically expressing transcription factors (TFs) – providing insight into cell differentiation and a powerful tool in both regenerative medicine and plant biotechnology. We are systematically exploring what can be achieved through TF-induced reprogramming in plants, opening new avenues to study plant cell fate and new approaches to shape plant form and function.
The “Forgotten” Generation: The far-reaching impact of the haploid gametophyte on plant genetics
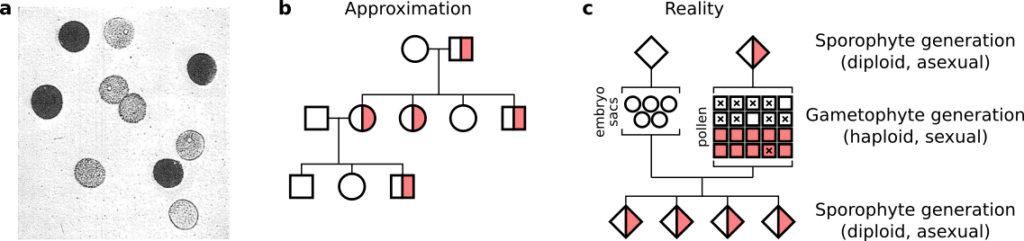
When we draw family trees in plant biology, we often focus on the relationship between diploid plants (Fig. 2b). This is an approximation. In reality, flowering plants alternate between a diploid generation and a multicellular haploid generation (Fig. 2c). One focus of the lab is to explore what happens when this approximation goes wrong.
Is there selection on pollen precursors during periods of abiotic stress? Pollen development is one of the most sensitive stages of the plant life cycle to abiotic stresses such as cold, heat, and drought. How much of this sensitivity is under sporophytic (diploid) vs gametophytic (haploid) genetic control? In other words, does abiotic stress select for favorable allele combinations in developing pollen? If so, environmental conditions would affect not only the quantity of pollen produced, but also the quality of the heritable material transmitted to the next diploid generation.
When does the pollen genome take over from its diploid parent? Most of the haploid life cycle occurs deep within developing flowers. How much of the haploid phase is actually under the control of the haploid genome? We found that there was a sharp and global transition from biallelic (diploid-derived) to monoallelic (haploid-derived) expression during the first pollen mitosis in maize (link). We are now exploring how the transition to haploid expression varies in other plant species. In collaboration with evolutionary biologists, we would like to learn: How much do plants modulate the extent of haploid selection they experience?
Can we make use of the haploid generation for more efficient genetics? Pollen provides a powerful yet underappreciated system for genetics and directed breeding, as it is produced with very high population numbers (>10 million pollen grains per diploid maize plant) and alleles that are recessive in the diploid phase are subject to direct selection in the haploid phase. We are developing high-throughput approaches to use each pollen grain as an independent genetic experiment, making it possible to test millions of gene combinations in every plant.
